Association Rule Mining and Gene Expression Analysis Uncover Links between Gastroesophageal Reflux disease and Diabetes Mellitus
Keywords:
Gastroesophageal Reflux Disease, Diabetes Mellitus, Association Rule MiningAbstract
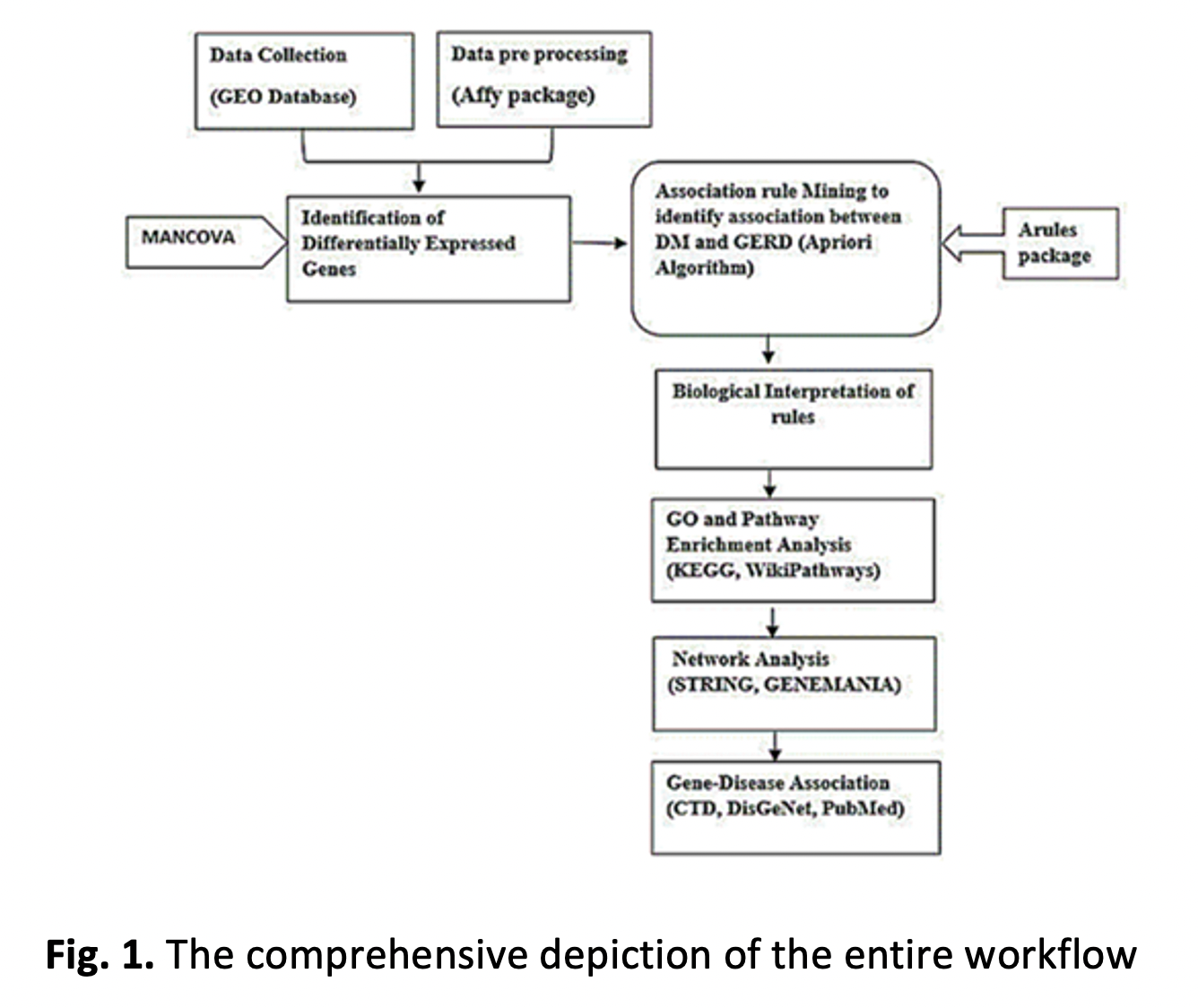
Gastroesophageal reflux disease and diabetes mellitus are prevalent chronic conditions with significant global health implications. Studies suggest a strong association between the two, with DM patients exhibiting a 61% higher risk of GERD due to factors such as delayed gastric emptying, autonomic neuropathy, and poor glycaemic control. Despite clinical evidence indicating a strong association between GERD and DM, the precise molecular mechanisms and shared genetic signatures underlying this comorbidity remain poorly understood. This knowledge gap limits the development of integrated diagnostic and therapeutic strategies for patients affected by both conditions. This study aims to determine shared gene expression profiles and signalling mechanisms that regulate the relationship between GERD and DM. Using gene expression datasets from the Gene Expression Omnibus (GEO), we identified 47 DEGs in GERD (GSE9768) and 17 in DM (GSE161355) by applying a rigorous statistical threshold (p < 0.002). To identify gene co-expression patterns, we employed an unsupervised machine learning technique, the Apriori algorithm of association rule mining, combined with Jaccard similarity, with minimum support (0.2) and confidence (0.8). This approach identified key genes including AXL, PTGER2, LGALS3, GBP1, and CHI3L1, which appeared in multiple rules, indicating their potential involvement in both diseases. Gene ontology and pathway enrichment analysis highlighted shared biological processes, particularly related to inflammation and prostaglandin signalling. Protein-protein interaction networks and gene co-expression analysis further supported the involvement of these genes in immune regulation and tissue remodelling. Gene-disease association analysis identified strong connections between the identified genes and both GERD and DM, offering potential therapeutic targets for further investigation. These findings suggest that common molecular mechanisms, particularly inflammatory pathways, underlie the coexistence of GERD and DM, and propose these genes as biomarkers for diagnosis and treatment strategies. Future studies should focus on experimental validation to confirm these results and explore potential therapeutic interventions.