Metagenomics Profiling of Microbial Community in Soil and Leachate Sample Isolated from a Landfill in Selangor, Malaysia
DOI:
https://doi.org/10.37934/sea.5.1.1935Keywords:
Metagenomics, microbial diversity, leachate, soil microbiome, functional pathwaysAbstract
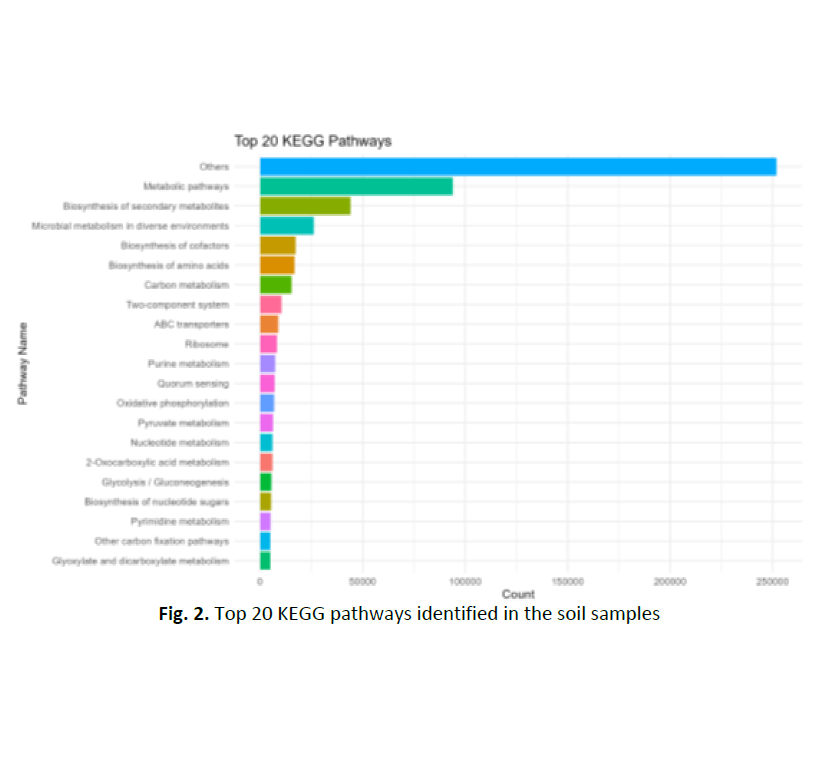
A landfill is a site for disposal of waste material by burying it in the ground. Understanding the impact of landfill ecosystems and studying the bioremediation methods require an understanding of their microbial diversity and functional ability. However, little is known about the microbial communities that found in landfill sites, especially in tropical areas. In order to further our knowledge of water quality and environmental sustainability, this study aims to outline the metabolic pathways and microbial community structure in soil and leachate samples from Jeram landfill. Sample of soil and leachate were carefully collected at the selected sites within the Jeram Landfill in Selangor, Malaysia. Metagenomic DNA was extracted and Illumina shotgun sequencing was performed for taxonomic and functional analysis. Taxonomic profiling showed that bacteria dominated in both soil (55.31%) and leachate (30.98%), whereas archaea were significantly more common in leachate (8.91%) than in soil (1.74%). Leachate contained a high percentage of unidentified sequences (59.60%), indicating the presence of a new or unknown microorganisms. The most prevalent species in the soil sample were Bacteroides graminisolvens, Methanosarcina barkeri, and Klebsiella pneumoniae. Methanogens like Methanoculleus bourgensis and Methanosarcina mazei, as well as Arcobacter species known for their roles in the nitrogen cycling, was dominant in leachate samples. KEGG pathway functional analysis revealed that metabolic pathway, biosynthesis of secondary metabolites, and carbon metabolism were the most common in both soil and leachate samples. However, leachate showed a significantly greater methane metabolism thus indicating the active anaerobic breakdown processes. These findings highlight how important microbial communities are to the breakdown of the organic matter, nitrogen cycling, and methane production which have a significant impact on the stability of landfills and the likelihood of water. This study enhances our knowledge of waste degrading processes and the implications for environmental sustainability and water resource management by providing insight into the metabolic processes and microbial diversity found in landfill sites. The findings provide an outline for further research on landfill impact assessment, bioremediation, and pollution control.